PCR is a method used to prepare billions of copies of specific DNA sequences. It is often necessary to have a larger number of copies of the specific DNA sequence than that found in a typical sample, to analyze DNA for its sequence or length. In addition, the PCR reaction is highly specific, meaning that it will only produce copies of a desired sequence from the template (sample) DNA. This specificity is ensured by the primers, which are designed to be complementary and anneal to specific regions on each side of the DNA region of interest (target region). These features make PCR a powerful technique to be used, for instance, in genotyping of markers, before sequencing, or in various DNA tests.
Figure 1. PCR consists of three steps: 1. Denaturation, 2. Annealing, and 3. Extension. The steps are repeated many times (often 30), producing billions of DNA copies of specific regions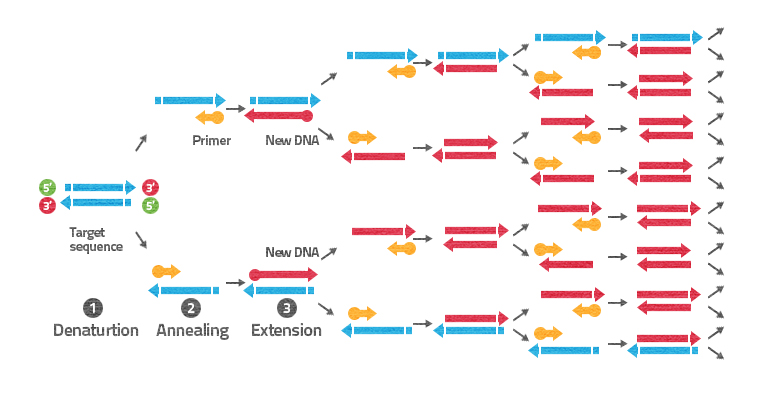
PCR steps
To prepare billions of DNA copies, many repeated cycles of DNA synthesis are performed in one PCR tube. Each cycle includes three distinct steps defined by the temperature (Figure 1). All cycles are performed without intervention in a PCR machine, which can change automatically the temperature to create the steps.
- Denaturation step (95ºC):在这么高的温度下,氢键及ng together the two DNA strands are broken, and the DNA strands fall apart. The single-stranded DNA is now available for copying.
- Annealing step (54ºC):At 54°C, short DNA pieces called primers bind at complementary sites of the template DNA. The primers define the target sequence, which is the specific region of DNA that will be copied. Annealing temperature is calculated from the primer composition (the number of nucleotides as well as number of guanine and cytosine). Normally, you would need to calculate the optimal annealing temperature for each primer. In this case, we consider 54°C as the annealing temperature for all the primers.
- Extension step (72ºC):At 72ºC, an enzyme called DNA polymerase is responsible for copying DNA. It recognizes the 3′ end of a primer bound to a template strand and starts copying the template DNA.
By the end of one cycle, parts of the initial DNA strands have been doubled in number. By the end of, e.g., 30 cycles, usually performed in PCR, at least 1 billion(230)copies of the target sequence will be present in the tube. For performing PCR, you need to add a thermostable DNA polymerase, nucleotides, primers, and DNA you want to use as your template.
PCR reagents
Several reagents are required to perform a PCR experiment;
- Primers:引物将以特定的DNA序列结合并标记DNA扩增的开始
- Nucleotides:Nucleotides are required to build the new DNA sequence
- Polymerase:Polymerase is an enzyme that assembles the nucleotides on the basis of the template sequence
- Template DNA:A template is required to be the basis of new DNA sequence amplification
When preparing for a PCR experiment, you must be extra careful for potential contaminations. PCR is a very powerful technique to amplify DNA. This means that if you have a tiny contamination (for example, DNA coming from other sample), this DNA can also be amplified, competing with the original template and destroying your experiment results. To prevent contamination, you need to always use gloves and work in a very clean environment (change the pipette tips when you are pipetting from a different container, tie your hair, and do not cough or sneeze around the PCR workbench).
Primer design for direct cloning
In order to create PCR products that can be cloned to plasmid directly, it is necessary to add restriction site at both ends of the PCR product. These restriction site addition can be done using a pair of primers with specific restriction site added at 5' end. Each primer contains a restriction site at 5' end and a hybridization site at 3' end. Consult therestriction site table选择正确的限制网站。选定的限制站点必须满足以下要求:
- Restriction enzymes that do not cut within RAD52 gene.
- 仅在目标质粒处切入MCS(多克隆位点)的限制酶。
- 具有相同最佳缓冲液和温度条件的限制酶。
Follow these step to create primer :
Forward primer
Choose 18-22 bp at the beginning of gene of interest sequence (make sure it satisfied the good primer requirements).
Add restriction site sequence at 5'end.
反向引物
Choose 18-22 bp at the end of gene of interest sequence (make sure it satisfied the good primer requirements).
Reverse complement the chosen sequence.
Add restriction sites sequence at 5' end.